Countries with highest deaths from covid
A lot of time was spent in 2020 looking at daily covid numbers, which was getting overwhelming. Having taken a break for a while, this article combines covid data and vaccination data with population data to present a view of the countries with the highest deaths from Covid-19 and the vaccination status in the country.
The Jupyter notebook is available on Github Countries-with-highest-deaths-from-covid-19-COVID-19 and it can be launched in a browser using Binder with this link Binder - Countries with highest deaths from COVID-19.
Load Covid data
As shown previously in Countries with highest confirmed cases of Covid-19 with Plotly the data is available from Johns Hopkins University who have made the data available on GitHub. More information about COVID-19 and the coronavirus is available from Coronavirus disease (COVID-19) advice for the public.
The latest data on deaths from Covid-19 is loaded directly from github.
1# Number of Death Cases - Global: time series data for deaths
2deaths_path = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv'
3
4deaths_17_df = pd.read_csv(deaths_path, usecols=['Country/Region', '12/17/21'])
5deaths_17_df = deaths_17_df.groupby("Country/Region").sum()
6deaths_17_df.rename_axis(None, axis=0, inplace=True)
7deaths_17_df
8
9"""
10 12/17/21
11Afghanistan 7332
12Albania 3158
13Algeria 6175
14Andorra 134
15Angola 1738
16Antigua and Barbuda 117
17Argentina 116892
18Armenia 7885
19Australia 2142
20Austria 13438
21"""
1Top_10 = deaths_17_df.sort_values(by = deaths_17_df.columns[-1], ascending = False).head(10)
2Top_10
3
4"""
5 12/17/21
6US 805823
7Brazil 617395
8India 476869
9Mexico 297356
10Russia 289292
11Peru 202076
12United Kingdom 147509
13Indonesia 143986
14Italy 135421
15Iran 130992
16"""
1bg_color = 'rgba(208,225,242,1.0)'
2latest_date = pd.to_datetime(deaths_17_df.columns[0])
3
4fig = go.Figure(
5 data = [go.Bar(x = list(Top_10.iloc[:, 0]),
6 y = list(Top_10.index),
7 hovertemplate = "%{y}: <br>Deaths: %{x:,}<extra></extra>",
8 orientation = "h")]
9)
10
11fig.update_layout(
12 # Set default font
13 font = dict(
14 family = "Droid Sans",
15 size = 14
16 ),
17 # Set figure title
18 title = dict(
19 text = f"<b>Countries with highest deaths from Covid [{latest_date.strftime('%b %d, %Y')}]</b>",
20 xref = 'container',
21 yref = 'container',
22 x = 0.5,
23 y = 0.87,
24 xanchor = 'center',
25 yanchor = 'middle',
26 font = dict(family = 'Droid Sans', size = 24)
27 ),
28 # set the plot bacground color
29 plot_bgcolor = bg_color,
30 # set the hover background color
31 hoverlabel = dict(
32 bgcolor = 'rgba(75,152,201,0.2)',
33 font_size = 16
34 ),
35 paper_bgcolor = bg_color,
36)
37
38fig.update_traces(marker_color = 'rgb(75,152,201)')
39fig['layout']['yaxis']['autorange'] = "reversed"
40fig.show()
Countries with highest deaths from COVID-19
Load Vaccination data
Covid-19 vaccination data is also available publicly on Github at COVID-19 data.
1# Number of Vaccines - Global: time series data for deaths
2vaccines_path = "https://raw.githubusercontent.com/govex/COVID-19/master/data_tables/vaccine_data/global_data/time_series_covid19_vaccine_global.csv"
3vaccines_df = pd.read_csv(vaccines_path)
4
5print(f"Shape of vaccines = {vaccines_df.shape}")
6"""
7Shape of vaccines = (129128, 8)
8"""
9
10print(vaccines_df.iloc[[0,1,2,-3,-2,-1], :])
11"""
12 Country_Region Date Doses_admin People_partially_vaccinated People_fully_vaccinated Report_Date_String UID Province_State
130 Canada 2020-12-14 5.0 0.0 0.0 2020-12-14 124.0 NaN
141 World 2020-12-14 5.0 0.0 0.0 2020-12-14 NaN NaN
152 Canada 2020-12-15 723.0 0.0 0.0 2020-12-15 124.0 NaN
16129125 Yemen 2021-12-17 786027.0 556652.0 366587.0 2021-12-18 887.0 NaN
17129126 Zambia 2021-12-17 1342989.0 806611.0 904201.0 2021-12-18 894.0 NaN
18129127 Zimbabwe 2021-12-17 7058474.0 4023289.0 3035185.0 2021-12-18 716.0 NaN
19"""
Look at vaccinations for a single Country in the most recent dates.
1vaccines_df[vaccines_df['Country_Region'] == 'Mexico'].tail(10)
2"""
3
4 Country_Region Date Doses_admin People_partially_vaccinated People_fully_vaccinated Report_Date_String UID Province_State
5123215 Mexico 2021-12-08 134370326.0 78816099.0 65543077.0 2021-12-09 484.0 NaN
6123849 Mexico 2021-12-09 135022230.0 78816099.0 65543077.0 2021-12-10 484.0 NaN
7124483 Mexico 2021-12-10 135481459.0 78816099.0 65543077.0 2021-12-11 484.0 NaN
8125117 Mexico 2021-12-11 136829147.0 80191983.0 66003384.0 2021-12-12 484.0 NaN
9125751 Mexico 2021-12-12 137169511.0 80388496.0 66150375.0 2021-12-13 484.0 NaN
10126385 Mexico 2021-12-13 137357032.0 80503005.0 66225140.0 2021-12-14 484.0 NaN
11127019 Mexico 2021-12-14 137357032.0 80503005.0 66225140.0 2021-12-15 484.0 NaN
12127651 Mexico 2021-12-15 137357032.0 81770394.0 66459570.0 2021-12-16 484.0 NaN
13128283 Mexico 2021-12-16 139820373.0 81770394.0 66459570.0 2021-12-17 484.0 NaN
14128915 Mexico 2021-12-17 139820373.0 81785946.0 66586509.0 2021-12-18 484.0 NaN
15"""
Look at vaccinations for a single date.
1vac_17_df = vaccines_df[vaccines_df['Date'] == '2021-12-17']
2print(vac_17_df.iloc[[0,1,2,4,-4,-3,-2,-1], :])
3
4"""
5 Country_Region Date Doses_admin People_partially_vaccinated People_fully_vaccinated Report_Date_String UID Province_State
6128496 Afghanistan 2021-12-17 5.228706e+06 4.397449e+06 3.566192e+06 2021-12-18 4.0 NaN
7128497 Albania 2021-12-17 2.226267e+06 1.111494e+06 1.001189e+06 2021-12-18 8.0 NaN
8128498 Algeria 2021-12-17 1.229306e+07 6.875003e+06 5.391232e+06 2021-12-18 12.0 NaN
9128500 Angola 2021-12-17 1.080528e+07 7.246240e+06 3.559041e+06 2021-12-18 24.0 NaN
10129124 World 2021-12-17 8.647128e+09 4.402818e+09 3.622553e+09 2021-12-18 NaN NaN
11129125 Yemen 2021-12-17 7.860270e+05 5.566520e+05 3.665870e+05 2021-12-18 887.0 NaN
12129126 Zambia 2021-12-17 1.342989e+06 8.066110e+05 9.042010e+05 2021-12-18 894.0 NaN
13129127 Zimbabwe 2021-12-17 7.058474e+06 4.023289e+06 3.035185e+06 2021-12-18 716.0 NaN
14"""
1fully_vac_17_df = vac_17_df.groupby("Country_Region").sum()[['People_fully_vaccinated']]
2fully_vac_17_df.rename_axis(None, axis=0, inplace=True)
3fully_vac_17_df.drop(['World', 'US (Aggregate)'], axis=0, inplace=True)
4
5top_10_vac = fully_vac_17_df.sort_values(by = fully_vac_17_df.columns[-1], ascending = False).head(10)
6top_10_vac
7
8"""
9 People_fully_vaccinated
10China 1.162488e+09
11India 5.330402e+08
12US 2.034792e+08
13Brazil 1.411751e+08
14Indonesia 1.052381e+08
15Japan 9.822253e+07
16Mexico 6.658651e+07
17Russia 6.207541e+07
18Pakistan 5.936147e+07
19Germany 5.817144e+07
20"""
Countries with highest vaccinations against COVID-19
Load population data
As detailed in Display COVID-19 numbers relative to population the population data is available from the World Bank data. This is available to download in csv, xml or Excel format. The Excel file contains three worksheets with the population data in the first sheet Data.
Access the excel file to determine the sheet names in the file.
1pop_excel_link = "https://api.worldbank.org/v2/en/indicator/SP.POP.TOTL?downloadformat=excel"
2
3pop_xl = pd.ExcelFile(pop_excel_link, engine = "xlrd")
4print(f"Worksheets = {pop_xl.sheet_names}") # see all sheet names
5
6"""
7Worksheets = ['Data', 'Metadata - Countries', 'Metadata - Indicators']
8"""
The first sheet *Data* contains the population information.
1# Load the excel worksheet into a dataframe
2pop_df = pd.read_excel(
3 pop_excel_link,
4 engine = "xlrd",
5 sheet_name = 'Data',
6 header = 3)
7
8print(pop_df.shape)
9"""
10(266, 65)
11"""
12
13print(pop_df.iloc[[0,1,2,-3,-2,-1], [0,1,2,-3,-2,-1]])
14"""
15 Country Name Country Code Indicator Name 2018 2019 2020
160 Aruba ABW Population, total 105846.0 106310.0 106766.0
171 Africa Eastern and Southern AFE Population, total 643090131.0 660046272.0 677243299.0
182 Afghanistan AFG Population, total 37171922.0 38041757.0 38928341.0
19263 South Africa ZAF Population, total 57792520.0 58558267.0 59308690.0
20264 Zambia ZMB Population, total 17351714.0 17861034.0 18383956.0
21265 Zimbabwe ZWE Population, total 14438812.0 14645473.0 14862927.0
22"""
This can be simplified to just read in the last column as we are interested in the latest population.
1# Load only the desired columns from the excel worksheet into a dataframe
2pop_2020_df = pd.read_excel(
3 pop_excel_link,
4 engine = "xlrd",
5 sheet_name = 'Data',
6 usecols = ['Country Name', 'Country Code', '2019'],
7 header = 3)
8
9pop_2020_df.set_index('Country Name', inplace=True)
10pop_2020_df.rename_axis(None, axis=0, inplace=True)
11print(pop_2020_df.shape)
12"""
13(266, 2)
14"""
15
16print(pop_2020_df.iloc[[0,1,2,3,4,5,-5,-4,-3,-2,-1], :])
17"""
18 Country Code 2019
19Aruba ABW 106310.0
20Africa Eastern and Southern AFE 660046272.0
21Afghanistan AFG 38041757.0
22Africa Western and Central AFW 446911598.0
23Angola AGO 31825299.0
24Albania ALB 2854191.0
25Kosovo XKX 1788878.0
26Yemen, Rep. YEM 29161922.0
27South Africa ZAF 58558267.0
28Zambia ZMB 17861034.0
29Zimbabwe ZWE 14645473.0
30"""
There are a number of regions that are aggregate of other countries. These are removed using the data in the second sheet in the excel file (Metadata - Countries).
1# Need to remove regions that are not countries
2metadata_df = pd.read_excel(
3 pop_excel_link,
4 engine = "xlrd",
5 sheet_name = 'Metadata - Countries',
6 header = 0)
7
8# metadata_df.isnan('Region')
9non_country_regions = list(metadata_df[metadata_df['Region'].isna()]['Country Code'])
10print(non_country_regions)
11
12"""
13['AFE', 'AFW', 'ARB', 'CEB', 'CSS', 'EAP', 'EAR', 'EAS', 'ECA', 'ECS', 'EMU', 'EUU', 'FCS', 'HIC', 'HPC', 'IBD', 'IBT', 'IDA', 'IDB', 'IDX', 'LAC', 'LCN', 'LDC', 'LIC', 'LMC', 'LMY', 'LTE', 'MEA', 'MIC', 'MNA', 'NAC', 'OED', 'OSS', 'PRE', 'PSS', 'PST', 'SAS', 'SSA', 'SSF', 'SST', 'TEA', 'TEC', 'TLA', 'TMN', 'TSA', 'TSS', 'UMC', 'WLD']
14"""
These are the top country populations in the world in 2020.
1# Remove non-countries from the dataframe
2pop_2020_df = pop_2020_df[~pop_2020_df['Country Code'].isin(non_country_regions)].copy()
3top_10_pop = pop_2020_df.sort_values(by = pop_2020_df.columns[-1], ascending = False).head(10)
4
5print(top_10_pop)
6"""
7 Country Code 2020
8China CHN 1.410929e+09
9India IND 1.380004e+09
10United States USA 3.294841e+08
11Indonesia IDN 2.735236e+08
12Pakistan PAK 2.208923e+08
13Brazil BRA 2.125594e+08
14Nigeria NGA 2.061396e+08
15Bangladesh BGD 1.646894e+08
16Russian Federation RUS 1.441041e+08
17Mexico MEX 1.289328e+08
18"""
Top 10 countries by population
Compare countries in the datasets
There are some discrepancies in the countries between the datasets.
1def extra_countries(df1, df2):
2 unmatched_covid_countries = sorted(
3 list(
4 df1[
5 ~df1.index
6 .isin(list(df2.index))
7 ].index))
8 return unmatched_covid_countries
9
10
11def print_list(data, columns=3):
12 # Pretty display for the list of countries
13 for i, x in enumerate(data):
14 print(f"{x.ljust(38, ' ')}", end='')
15 if i%columns==(columns-1):
16 print()
1extra = extra_countries(deaths_17_df, pop_2020_df)
2print_list(extra)
3
4"""
5Bahamas Brunei Burma
6Congo (Brazzaville) Congo (Kinshasa) Czechia
7Diamond Princess Egypt Gambia
8Holy See Iran Korea, South
9Kyrgyzstan Laos MS Zaandam
10Micronesia Russia Saint Kitts and Nevis
11Saint Lucia Saint Vincent and the Grenadines Slovakia
12Summer Olympics 2020 Syria Taiwan*
13US Venezuela Yemen
14"""
1extra = extra_countries(pop_2020_df, deaths_17_df)
2print_list(extra)
3
4"""
5American Samoa Aruba Bahamas, The
6Bermuda British Virgin Islands Brunei Darussalam
7Cayman Islands Channel Islands Congo, Dem. Rep.
8Congo, Rep. Curacao Czech Republic
9Egypt, Arab Rep. Faroe Islands French Polynesia
10Gambia, The Gibraltar Greenland
11Guam Hong Kong SAR, China Iran, Islamic Rep.
12Isle of Man Korea, Dem. People's Rep. Korea, Rep.
13Kyrgyz Republic Lao PDR Macao SAR, China
14Micronesia, Fed. Sts. Myanmar Nauru
15New Caledonia Northern Mariana Islands Not classified
16Puerto Rico Russian Federation Sint Maarten (Dutch part)
17Slovak Republic St. Kitts and Nevis St. Lucia
18St. Martin (French part) St. Vincent and the Grenadines Syrian Arab Republic
19Turkmenistan Turks and Caicos Islands Tuvalu
20United States Venezuela, RB Virgin Islands (U.S.)
21Yemen, Rep.
22"""
1extra = extra_countries(deaths_17_df, fully_vac_17_df)
2print_list(extra)
3
4"""
5Diamond Princess Eritrea Holy See
6MS Zaandam Summer Olympics 2020 Tonga
7"""
1extra = extra_countries(fully_vac_17_df, deaths_17_df)
2print_list(extra)
3"""
4empty list
5"""
1extra = extra_countries(fully_vac_17_df, pop_2020_df)
2print_list(extra)
3
4"""
5Bahamas Brunei Burma
6Congo (Brazzaville) Congo (Kinshasa) Czechia
7Egypt Gambia Iran
8Korea, South Kyrgyzstan Laos
9Micronesia Russia Saint Kitts and Nevis
10Saint Lucia Saint Vincent and the Grenadines Slovakia
11Syria Taiwan* US
12Venezuela Yemen
13"""
1extra = extra_countries(pop_2020_df, fully_vac_17_df)
2print_list(extra)
3
4"""
5American Samoa Aruba Bahamas, The
6Bermuda British Virgin Islands Brunei Darussalam
7Cayman Islands Channel Islands Congo, Dem. Rep.
8Congo, Rep. Curacao Czech Republic
9Egypt, Arab Rep. Eritrea Faroe Islands
10French Polynesia Gambia, The Gibraltar
11Greenland Guam Hong Kong SAR, China
12Iran, Islamic Rep. Isle of Man Korea, Dem. People's Rep.
13Korea, Rep. Kyrgyz Republic Lao PDR
14Macao SAR, China Micronesia, Fed. Sts. Myanmar
15Nauru New Caledonia Northern Mariana Islands
16Not classified Puerto Rico Russian Federation
17Sint Maarten (Dutch part) Slovak Republic St. Kitts and Nevis
18St. Lucia St. Martin (French part) St. Vincent and the Grenadines
19Syrian Arab Republic Tonga Turkmenistan
20Turks and Caicos Islands Tuvalu United States
21Venezuela, RB Virgin Islands (U.S.) Yemen, Rep.
22"""
Create a mapping between the country names in the datasets.
1unmatched_covid_countries = extra_countries(deaths_17_df, pop_2020_df)
2
3# remove cruise ships and disputed teritories that don't have any equivalent country in population dataset
4unmatched_covid_countries.remove('Diamond Princess')
5unmatched_covid_countries.remove('MS Zaandam')
6unmatched_covid_countries.remove('Holy See')
7unmatched_covid_countries.remove('Taiwan*')
8unmatched_covid_countries.remove('Summer Olympics 2020')
9
10# Manually map these countries to the country names used in COVID-19 dataset
11map_pop = ['Bahamas, The', 'Brunei Darussalam', 'Myanmar', 'Congo, Rep.', 'Congo, Dem. Rep.',
12 'Czech Republic', 'Egypt, Arab Rep.', 'Gambia, The', 'Iran, Islamic Rep.',
13 'Korea, Rep.', 'Kyrgyz Republic', 'Lao PDR', 'Micronesia, Fed. Sts.',
14 'Russian Federation', 'St. Kitts and Nevis', 'St. Lucia', 'St. Vincent and the Grenadines',
15 'Slovak Republic', 'Syrian Arab Republic', 'United States', 'Venezuela, RB', 'Yemen, Rep.'
16 ]
17
18country_map = {}
19for c,p in zip(unmatched_covid_countries, map_pop):
20 country_map[p] = c
Mapping of mis-matched country names between Covid-data and Population data:
| COVID-19 Country Name | Population Country Name |
|---|---|
| Bahamas | Bahamas, The |
| Brunei | Brunei Darussalam |
| Burma | Myanmar |
| Congo (Brazzaville) | Congo, Rep. |
| Congo (Kinshasa) | Congo, Dem. Rep. |
| Czechia | Czech Republic |
| Egypt | Egypt, Arab Rep. |
| Gambia | Gambia, The |
| Iran | Iran, Islamic Rep. |
| Korea, South | Korea, Rep. |
| Kyrgyzstan | Kyrgyz Republic |
| Laos | Lao PDR |
| Micronesia | Micronesia, Fed. Sts. |
| Russia | Russian Federation |
| Saint Kitts and Nevis | St. Kitts and Nevis |
| Saint Lucia | St. Lucia |
| Saint Vincent and the Grenadines | St. Vincent and the Grenadines |
| Slovakia | Slovak Republic |
| Syria | Syrian Arab Republic |
| US | United States |
| Venezuela | Venezuela, RB |
| Yemen | Yemen, Rep. |
Add a field to the population data to contain the country name.
1# Add the covid country to the population dataset
2pop_2020_df['Covid_Country'] = pop_2020_df.apply(
3 lambda x: country_map[x.name]
4 if x.name in country_map
5 else x.name,
6 axis = 1)
7
8print(pop_2020_df.iloc[[1,2,3,4,-4,-3,-2,-1], :])
9
10"""
11 Country Code 2020 Covid_Country
12Afghanistan AFG 38928341.0 Afghanistan
13Angola AGO 32866268.0 Angola
14Albania ALB 2837743.0 Albania
15Andorra AND 77265.0 Andorra
16Yemen, Rep. YEM 29825968.0 Yemen
17South Africa ZAF 59308690.0 South Africa
18Zambia ZMB 18383956.0 Zambia
19Zimbabwe ZWE 14862927.0 Zimbabwe
20"""
Confirm that there are no unmatched countries in the Covid datasets.
1print(list(deaths_17_df[
2 ~deaths_17_df.index
3 .isin(list(pop_2020_df['Covid_Country']))
4].index))
5
6"""
7['Diamond Princess', 'Holy See', 'MS Zaandam', 'Summer Olympics 2020', 'Taiwan*']
8"""
9
10print(list(fully_vac_17_df[
11 ~fully_vac_17_df.index
12 .isin(list(pop_2020_df['Covid_Country']))
13].index))
14
15"""
16['Taiwan*']
17"""
Merge datasets on country
A new dataframe is created with the data for the deaths from covid-19 and the vaccinations as well as the population for each country.
1covid_pop_df = deaths_17_df.merge(
2 fully_vac_17_df,
3 how='left',
4 left_index = True,
5 right_on = fully_vac_17_df.index
6).merge (
7 pop_2020_df,
8 how='left',
9 left_on = covid_df.index,
10 right_on = 'Covid_Country'
11)
12
13covid_pop_df.set_index('key_0', inplace=True)
14covid_pop_df.rename_axis(None, axis=0, inplace=True)
15
16# # Rename column names for deaths and population
17covid_pop_df = covid_pop_df.rename(columns={'12/17/21': 'Deaths'})
18covid_pop_df = covid_pop_df.rename(columns={'2020': 'Population'})
19
20# # Remove unneeded columns
21covid_pop_df.drop(['Country Code', 'Covid_Country'], axis=1, inplace=True)
22
23print(covid_pop_df.iloc[[1,2,3,4,-4,-3,-2,-1], :])
24
25"""
26 Deaths People_fully_vaccinated Population
27Albania 3158 1001189.0 2837743.0
28Algeria 6175 5391232.0 43851043.0
29Andorra 134 50551.0 77265.0
30Angola 1738 3559041.0 32866268.0
31West Bank and Gaza 4855 0.0 4803269.0
32Yemen 1974 366587.0 29825968.0
33Zambia 3674 904201.0 18383956.0
34Zimbabwe 4779 3035185.0 14862927.0
35"""
Overview of Countries with highest deaths from covid per capita
The number of people vaccinated is calculated per 1,000 of the population and the number of people died is calculated per 100,000 of the population. The top countries with the highest deaths per capita and those with the highest vaccinations are shown for countries with populations greater than 1 million.
1covid_pop_df['deaths_100k'] = covid_pop_df.apply(lambda x: (x.Deaths / x.Population) * 100000, axis = 1)
2
3covid_pop_df['vac_100'] = covid_pop_df.apply(lambda x: (x.People_fully_vaccinated / x.Population) * 100, axis = 1)
4
5print(covid_pop_df.iloc[[1,2,3,4,-4,-3,-2,-1], :])
6
7"""
8 Deaths People_fully_vaccinated Population deaths_100k vac_100
9Albania 3158 1001189.0 2837743.0 111.285624 35.281172
10Algeria 6175 5391232.0 43851043.0 14.081763 12.294421
11Andorra 134 50551.0 77265.0 173.429108 65.425484
12Angola 1738 3559041.0 32866268.0 5.288097 10.828857
13West Bank and Gaza 4855 0.0 4803269.0 101.076996 0.000000
14Yemen 1974 366587.0 29825968.0 6.618394 1.229087
15Zambia 3674 904201.0 18383956.0 19.984817 4.918425
16Zimbabwe 4779 3035185.0 14862927.0 32.153828 20.421179
17"""
1top_10 = (covid_pop_df[covid_pop_df['Population'] > 1000000]
2 .sort_values(by = ['deaths_100k'], ascending = False)
3 .head(20))
4
5bg_color = 'rgba(208,225,242,1.0)'
6
7fig = go.Figure(
8 data = [go.Bar(x = list(top_10["deaths_100k"]),
9 y = list(top_10.index),
10 hovertemplate = "%{y}: <br>Deaths per 100K: %{x:.0f}<extra></extra>",
11 orientation = "h")]
12)
13
14fig.update_layout(
15 # Set default font
16 font = dict(
17 family = "Droid Sans",
18 size = 14
19 ),
20 # Set figure title
21 title = dict(
22 text = f"<b>Countries with highest deaths from Covid-19 per 100K of population [Dec 17, 2020]</b>",
23 xref = 'container',
24 yref = 'container',
25 x = 0.5,
26 y = 0.92,
27 xanchor = 'center',
28 yanchor = 'middle',
29 font = dict(family = 'Droid Sans', size = 24)
30 ),
31 # set the plot bacground color
32 plot_bgcolor = bg_color,
33 # set the hover background color
34 hoverlabel = dict(
35 bgcolor = 'rgba(75,152,201,0.2)',
36 font_size = 16
37 ),
38 paper_bgcolor = bg_color,
39)
40
41fig.update_traces(marker_color = 'rgb(75,152,201)')
42fig['layout']['yaxis']['autorange'] = "reversed"
43fig.show()
Countries with highest deaths from Covid-19 per 100K of population
These are the countries with populations over 1 million with the lowest number of deaths from covid-19 per capita.
Countries with lowest deaths from Covid-19 per 100K of population
These are the countries with populations over 1 million with the highest percent of the population vaccinated against covid-19.
Countries with lowest deaths from Covid-19 per 100K of population
Vaccination status in countries with highest deaths
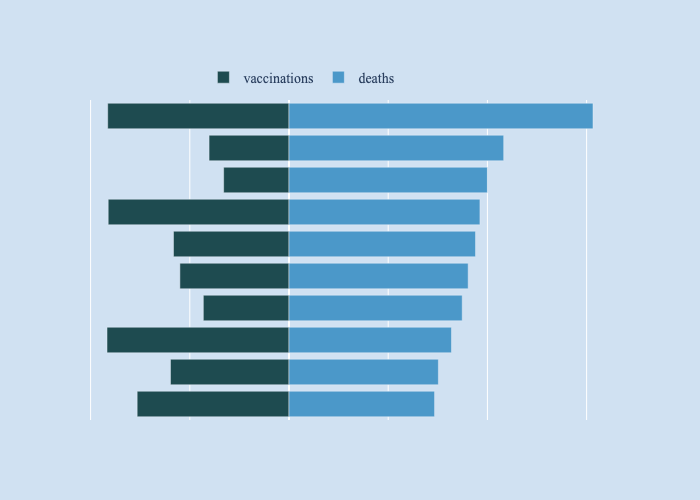
A bar plot in relative mode is used to show percent vaccinated with number of deaths in countries with the highest deaths from cvid-19.
1top_20 = (covid_pop_df[covid_pop_df['Population'] > 1000000]
2 .sort_values(by = ['deaths_100k'], ascending = False)
3 .head(20))
4
5neg_vac = [(-1 * x) for x in top_20.vac_100]
6
7bg_color = 'rgba(208,225,242,1.0)'
8
9fig = go.Figure()
10fig.add_trace(go.Bar(x = [v*6 for v in neg_vac],
11 y = list(top_20.index),
12 marker_color = 'rgb(30,75,80)',
13 name = 'vaccinations',
14 customdata = [(v * -1) for v in neg_vac],
15 hovertemplate = "%{y}: <br>Vaccinated: %{customdata:.0f}%<extra></extra>",
16 orientation = "h"))
17fig.add_trace(go.Bar(x = list(top_20["deaths_100k"]),
18 y = list(top_20.index),
19 marker_color = 'rgb(75,152,201)',
20 name = 'deaths',
21 hovertemplate = "%{y}: <br>Deaths per 100K: %{x:.0f}<extra></extra>",
22 orientation = "h"))
23
24
25fig.update_layout(
26 # Set default font
27 font = dict(
28 family = "Droid Sans",
29 size = 14
30 ),
31 # Set figure title
32 title = dict(
33 text = f"<b>Countries with highest deaths from Covid-19 per 100K of population [Dec 17, 2020]</b>",
34 xref = 'container',
35 yref = 'container',
36 x = 0.5,
37 y = 0.95,
38 xanchor = 'center',
39 yanchor = 'middle',
40 font = dict(family = 'Droid Sans', size = 24)
41 ),
42 # set x-axis
43 xaxis = dict(
44 title = '',
45 showticklabels = False,
46 ),
47
48 legend=dict(
49 orientation="h",
50 yanchor="bottom",
51 y=1.02,
52 xanchor="center",
53 x=0.41
54 ),
55
56 # set the plot bacground color
57 plot_bgcolor = bg_color,
58 # set the hover background color
59 hoverlabel = dict(
60 bgcolor = 'rgba(75,152,201,0.2)',
61 font_size = 16
62 ),
63 paper_bgcolor = bg_color,
64)
65
66fig.update_layout(barmode='relative')
67fig['layout']['yaxis']['autorange'] = "reversed"
68fig.show()
Countries with highest deaths from Covid-19 per 100K of population with percentage vaccinated
Conclusion
The covid-19 pandemic is far from over and their continues to be a number of Country status for Covid and Vaccinations. There is a trend that those countries with higher percentage of the population vaccinated have lower deaths per capita, but there are exceptions to this.