Pandas - Load data from Excel file and Display Chart
Pandas is a Python package that provides fast data manipulation and analysis of
structured data. It has grown in popularity recently with the rise of data analytics
and machine learning. The source code for pandas is available on GitHub, but it is
more common to install the binary package using something like pip install pandas.
A core component of pandas is the dataframe that provides two-dimensional
table-like structure to store and manipulate data.
Pandas makes working with structured data easy using the flexible data structures provided. It has become an essential tool for doing practical data analysis in Python. Using Pandas in Jupyter notebooks is a very efficient way to load data and perform some high-level analysis and then visualize the data using Matplotlib.
Under-five mortality rate
I came across a book from Unicef called "state of the world's children" in 1995. This reports on key issues affecting children in various countries around the world and includes a large set of statistics in tabular format, which I found fascinating. Unicef works to improve child welfare throughout the world and continues to produce numerous reports including the state of the word's children. Unicef makes available a large amount of datasets for public consumption at Unicef Datasets. This article uses Under 5 Mortality Rate dataset.
Under-five mortality rate:
is the probability of dying between birth and exactly 5 years of age, expressed per 1,000 live births.
Pandas functions used in this article |
|
|---|---|
|
|
Python modules used in this article
1# import modules
2import matplotlib.pyplot as plt
3import shutil
4import pandas as pd
5import numpy as np
6import requests
7import os
Download the excel file
Use the following function to download any file to a temporary location. It is always better to copy data locally and then work with the local files.
1def download_file(url):
2 '''Download a file from url to local temp directory.
3
4 Keyword arguments:
5 url -- A fully qualified url file path
6
7 return: The fully qualified path to the downloaded file
8 '''
9 local_dir = '/tmp/data/'
10 if not os.path.exists(local_dir):
11 os.makedirs(local_dir)
12
13 local_file = local_dir + url.split('/')[-1]
14 r = requests.get(url, stream=True)
15 with open(local_file, 'wb') as f:
16 for chunk in r.iter_content(chunk_size=512*1024):
17 if chunk:
18 f.write(chunk)
19 return local_file
This function is used by passing in the url to the Excel file on under-five mortality rate.
1u5mr_url = 'https://data.unicef.org/wp-content/uploads/2020/09/Under-five-mortality-rate_2020.xlsx'
2u5mr_file = download_file(u5mr_url)
3
4print(f"local file = {u5mr_file}")
5# local file = /tmp/data/Under-five-mortality-rate_2020.xlsx
List the worksheets in an excel file
Loading an excel file into a Pandas dataframe is very straight forward with
read_excel() and it will default to loading the first worksheet in the Excel
file. It is also possible to find out what sheets are in an Excel file with
the following code. This shows that the file contains six worksheets and the
first one is the country estimates needed.
1u5mr_xl = pd.ExcelFile(u5mr_file)
2u5mr_xl.sheet_names
3
4
5# output
6'''
7['Country estimates (both sexes)',
8 'Regional & global (both sexes)',
9 'Country estimates (Female)',
10 'Regional & global (Female)',
11 'Country estimates (Male)',
12 'Regional & global (Male)']
13 '''
Load excel worksheet into dataframe
First load the first worksheet into a dataframe and examine the data. Examine the
data in the dataframe using shape and head. The shape of the data is (601, 73),
which is 601 rows of data by 73 columns. The head of the dataframe shows the columns
do not have any names and there seems to be a lot of blank rows at the top.
DataFrame.head function returns the first n rows of the dataframe, the default is 5. The first 5 rows appear largely blank except for file information in the first column. The data can be seen to start at row index 13 - when the head function is passed 20 for the first 20 rows.
Displaying all 73 columns can be somewhat unwieldy so the DataFrame.iloc function
is used to display the first 20 rows for the first 5 columns. iloc is an integer
based indexing mechanism for selecting rows and columns by position. The tail
function can be used in a similar way to the head function except to display the last
n rows of the dataframe. iloc is used here as well to reduce the number of columns to
display.
1u5mr_df = pd.read_excel(u5mr_file)
2
3# OR
4# u5mr_df = pd.read_excel(u5mr_file, sheet_name = 'Country estimates (both sexes)')
5
6print(u5mr_df.shape)
7"""
8(601, 73)
9"""
10
11u5mr_df.head()
12u5mr_df.head(20)
13
14print(u5mr_df.iloc[:20, 0:5])
15"""
16 Unnamed: 0 Unnamed: 1 UNICEF GLOBAL DATABASES [data.unicef.org] Unnamed: 3 Unnamed: 4
170 NaN NaN NaN NaN NaN
181 NaN NaN NaN NaN NaN
192 Child Mortality Estimates NaN NaN NaN NaN
203 Country-specific Under-five mortality rate, bo... NaN NaN NaN NaN
214 NaN NaN NaN NaN NaN
225 Estimates generated by the UN Inter-agency Gro... NaN NaN NaN NaN
236 Downloaded from http://data.unicef.org NaN NaN NaN NaN
247 NaN NaN NaN NaN NaN
258 Prepared by the Data and Analytics Section; Di... NaN NaN NaN NaN
269 Contact us: data@unicef.org NaN NaN NaN NaN
2710 NaN NaN NaN NaN NaN
2811 Last update: 9 September 2020 NaN NaN NaN NaN
2912 NaN NaN NaN NaN NaN
3013 ISO.Code Country.Name Uncertainty.Bounds* 1950.5 1951.5
3114 AFG Afghanistan Lower NaN NaN
3215 AFG Afghanistan Median NaN NaN
3316 AFG Afghanistan Upper NaN NaN
3417 ALB Albania Lower NaN NaN
3518 ALB Albania Median NaN NaN
3619 ALB Albania Upper NaN NaN
37"""
38
39
40u5mr_df.tail()
41print(u5mr_df.iloc[-5:, 0:5])
42"""
43 Unnamed: 0 Unnamed: 1 UNICEF GLOBAL DATABASES [data.unicef.org] Unnamed: 3 Unnamed: 4
44596 ZWE Zimbabwe Lower NaN NaN
45597 ZWE Zimbabwe Median NaN NaN
46598 ZWE Zimbabwe Upper NaN NaN
47599 NaN NaN NaN NaN NaN
48600 NaN * Lower and Upper refer to the lower bound and... NaN NaN NaN
49"""
It can be seen from the output above that the header row for the countries start at index 13. The data is reloaded from the excel file with an additional parameter specifying the header row. This could also be done using the skiprows. It also shows that the last two rows are there to contain a footnote.
Reload the excel file with the following code and review the data.
1u5mr_df = pd.read_excel(
2 u5mr_file,
3 sheet_name = 'Country estimates (both sexes)',
4 header = 14)
5
6print(u5mr_df.shape)
7"""
8(587, 73)
9"""
10
11print(u5mr_df.iloc[:10, 0:8])
12"""
13 ISO.Code Country.Name Uncertainty.Bounds* 1950.5 1951.5 1952.5 1953.5 1954.5
140 AFG Afghanistan Lower NaN NaN NaN NaN NaN
151 AFG Afghanistan Median NaN NaN NaN NaN NaN
162 AFG Afghanistan Upper NaN NaN NaN NaN NaN
173 ALB Albania Lower NaN NaN NaN NaN NaN
184 ALB Albania Median NaN NaN NaN NaN NaN
195 ALB Albania Upper NaN NaN NaN NaN NaN
206 DZA Algeria Lower NaN NaN NaN NaN 181.095341
217 DZA Algeria Median NaN NaN NaN NaN 242.618134
228 DZA Algeria Upper NaN NaN NaN NaN 323.057628
239 AND Andorra Lower NaN NaN NaN NaN NaN
24"""
25
26# Review columns on the right
27print(u5mr_df.iloc[:10, -7:])
28"""
29 2013.5 2014.5 2015.5 2016.5 2017.5 2018.5 2019.5
300 68.481212 64.526394 60.716842 57.068054 53.744798 50.640922 47.442070
311 76.830462 73.580267 70.444802 67.572190 64.940759 62.541196 60.269399
322 86.077559 83.378062 80.863431 78.646207 76.832873 75.405564 74.616041
333 10.072106 9.549495 9.225420 9.075629 9.044506 9.067390 9.068154
344 10.430484 9.896397 9.565251 9.419110 9.418052 9.525133 9.682407
355 10.821932 10.272281 9.936220 9.795035 9.826251 10.020056 10.348964
366 25.265117 24.944603 24.613434 24.245765 23.801969 23.262592 22.513380
377 25.784460 25.478560 25.177142 24.792098 24.319482 23.805926 23.256168
388 26.310524 26.023294 25.746094 25.339485 24.843878 24.345820 24.025946
399 1.566198 1.384200 1.232419 1.098862 0.985541 0.910653 0.847791
40"""
Rename the columns to contain the Year
Drop the last two rows that are there to include the footnote.
1# Drop the last two rows
2print(u5mr_df.shape)
3"""
4(587, 73)
5"""
6
7u5mr_df.drop(u5mr_df.tail(2).index, inplace = True)
8
9print(u5mr_df.shape)
10"""
11(585, 73)
12"""
13
14u5mr_df.tail()
15print(u5mr_df.iloc[-5:, 0:7])
16"""
17 ISO.Code Country.Name Uncertainty.Bounds* 1950.5 1951.5 1952.5 1953.5
18580 ZMB Zambia Median NaN NaN NaN 234.418232
19581 ZMB Zambia Upper NaN NaN NaN 308.531403
20582 ZWE Zimbabwe Lower NaN NaN NaN NaN
21583 ZWE Zimbabwe Median NaN NaN NaN NaN
22584 ZWE Zimbabwe Upper NaN NaN NaN NaN
23"""
The year headings contain 0.5 such as 2019.5 probably to indicate a mean value for
the entire year. Use columns field of the dataframe to rename these to just the
full year. u5mr_df.columns is a list of all the columns in the dataframe and
a list comprehension is used to remove the ".5" if it is there.
The asterisk (*) is also removed from the 'Uncertainty.Bounds*' column name.
1print(u5mr_df.columns)
2
3"""
4Index(['ISO.Code', 'Country.Name', 'Uncertainty.Bounds*', '1950.5', '1951.5',
5 '1952.5', '1953.5', '1954.5', '1955.5', '1956.5', '1957.5', '1958.5',
6 '1959.5', '1960.5', '1961.5', '1962.5', '1963.5', '1964.5', '1965.5',
7 '1966.5', '1967.5', '1968.5', '1969.5', '1970.5', '1971.5', '1972.5',
8 '1973.5', '1974.5', '1975.5', '1976.5', '1977.5', '1978.5', '1979.5',
9 '1980.5', '1981.5', '1982.5', '1983.5', '1984.5', '1985.5', '1986.5',
10 '1987.5', '1988.5', '1989.5', '1990.5', '1991.5', '1992.5', '1993.5',
11 '1994.5', '1995.5', '1996.5', '1997.5', '1998.5', '1999.5', '2000.5',
12 '2001.5', '2002.5', '2003.5', '2004.5', '2005.5', '2006.5', '2007.5',
13 '2008.5', '2009.5', '2010.5', '2011.5', '2012.5', '2013.5', '2014.5',
14 '2015.5', '2016.5', '2017.5', '2018.5', '2019.5'],
15 dtype='object')
16"""
17
18u5mr_df.columns = [x[:-2] if x.endswith('.5') else x for x in u5mr_df.columns]
19
20u5mr_df = u5mr_df.rename(columns={'Uncertainty.Bounds*': 'Uncertainty.Bounds'})
21
22print(u5mr_df.columns)
23"""
24Index(['ISO.Code', 'Country.Name', 'Uncertainty.Bounds', '1950', '1951',
25 '1952', '1953', '1954', '1955', '1956', '1957', '1958', '1959', '1960',
26 '1961', '1962', '1963', '1964', '1965', '1966', '1967', '1968', '1969',
27 '1970', '1971', '1972', '1973', '1974', '1975', '1976', '1977', '1978',
28 '1979', '1980', '1981', '1982', '1983', '1984', '1985', '1986', '1987',
29 '1988', '1989', '1990', '1991', '1992', '1993', '1994', '1995', '1996',
30 '1997', '1998', '1999', '2000', '2001', '2002', '2003', '2004', '2005',
31 '2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014',
32 '2015', '2016', '2017', '2018', '2019'],
33 dtype='object')
34"""
Filter to the Median values
The data has three values for each country the Lower and Upper bounds and the Median. Create a copy of the dataframe with just the Median values, so there is only one number for each country.
1u5mr_med_df = u5mr_df[u5mr_df['Uncertainty.Bounds'] == 'Median']
2print(u5mr_med_df.shape)
3"""
4(195, 73)
5"""
6
7u5mr_med_df.head()
8print(u5mr_med_df.iloc[:10, :10])
9"""
10 ISO.Code Country.Name Uncertainty.Bounds 1950 1951 1952 1953 1954 1955 1956
111 AFG Afghanistan Median NaN NaN NaN NaN NaN NaN NaN
124 ALB Albania Median NaN NaN NaN NaN NaN NaN NaN
137 DZA Algeria Median NaN NaN NaN NaN 242.618134 242.066662 241.601453
1410 AND Andorra Median NaN NaN NaN NaN NaN NaN NaN
1513 AGO Angola Median NaN NaN NaN NaN NaN NaN NaN
1616 ATG Antigua and Barbuda Median 119.066130 116.624640 114.074405 111.406742 108.602334 105.661011 102.585410
1719 ARG Argentina Median NaN NaN NaN NaN NaN NaN NaN
1822 ARM Armenia Median NaN NaN NaN NaN NaN NaN NaN
1925 AUS Australia Median 31.584087 30.966060 30.261701 29.496194 28.711058 27.961205 27.265216
2028 AUT Austria Median 83.482005 73.820239 66.122978 60.263643 56.225511 53.524855 51.536432
21"""
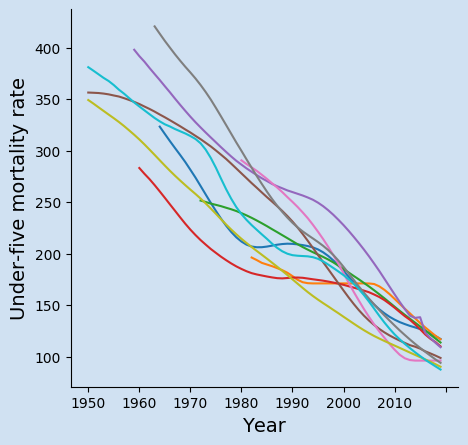
Get data for the top 10 countries in the latest year
The data is sorted by the values in the last column and the number of rows reduced to ten to reduce the data to the top 10 countries.
1u5mr_med_df.sort_values(by=u5mr_df.columns[-1], ascending=False).head(10)
2
3highest_df = u5mr_med_df.sort_values(by=u5mr_df.columns[-1], ascending=False).head(10)
4print(highest_df.iloc[:, [0,1,-5,-4,-3,-2,-1]])
5"""
6 ISO.Code Country.Name 2015 2016 2017 2018 2019
7376 NGA Nigeria 126.833543 125.040170 122.798947 120.037728 117.202078
8481 SOM Somalia 132.720296 128.428442 124.393442 120.326222 116.972096
9100 TCD Chad 129.406257 125.604878 121.508350 117.664883 113.790418
1097 CAF Central African Republic 126.714463 121.593307 117.470988 114.222380 110.053912
11466 SLE Sierra Leone 138.342774 123.232842 118.222181 113.544491 109.236528
12214 GIN Guinea 107.852191 105.241523 103.226528 101.095543 98.802973
13487 SSD South Sudan 96.229299 96.229299 96.229299 96.229299 96.229299
14316 MLI Mali 108.586504 104.766458 101.062422 97.410316 94.035418
1555 BEN Benin 99.592931 97.416395 95.133079 92.773521 90.286429
1679 BFA Burkina Faso 100.671924 97.034768 93.862909 90.682138 87.542426
17"""
Create a chart for the top 10 countries in the latest year
Create a line chart using Matplotlib to show the changes in the under-five mortality rate over time. The data needs to be transposed and tidyied up a bit in order to create a line chart using year as the x axis values.
The following steps are done to prepare the data for the chart.
- Transpose the data so year can be used as the x-axis
- Set the Country.Name as the heading for the columns
- Rename the Country.Name column to Year
- Drop the rows that do not contain u5mr data
1# Need to transpose the dataframe to use year as the x-axis
2top_10_df = u5mr_med_df.sort_values(by=u5mr_df.columns[-1], ascending=False).head(10).T
3top_10_df.reset_index(drop=False,inplace=True)
4print(f"shape = {top_10_df.shape}")
5top_10_df.head()
6
7# Set the Country.Name as the heading for the columns
8top_10_df.columns = top_10_df.iloc[np.where(top_10_df['index'] == 'Country.Name')[0][0]]
9print(f"shape = {top_10_df.shape}")
10top_10_df.head()
11
12# Rename the Country.Name column to Year
13top_10_df = top_10_df.rename(columns={'Country.Name': 'Year'})
14print(f"shape = {top_10_df.shape}")
15top_10_df.head()
16
17# Drop the rows that do not contain u5mr data
18top_10_df = top_10_df[top_10_df['Year'].isin(['ISO.Code', 'Country.Name', 'Uncertainty.Bounds']) == False]
19top_10_df.reset_index(drop = True, inplace = True)
20print(f"shape = {top_10_df.shape}")
21top_10_df.head()
22
23print(top_10_df.tail())
24"""
251 Year Nigeria Somalia Chad Central African Republic Sierra Leone Guinea South Sudan Mali Benin Burkina Faso
2665 2015 126.834 132.72 129.406 126.714 138.343 107.852 96.2293 108.587 99.5929 100.672
2766 2016 125.04 128.428 125.605 121.593 123.233 105.242 96.2293 104.766 97.4164 97.0348
2867 2017 122.799 124.393 121.508 117.471 118.222 103.227 96.2293 101.062 95.1331 93.8629
2968 2018 120.038 120.326 117.665 114.222 113.544 101.096 96.2293 97.4103 92.7735 90.6821
3069 2019 117.202 116.972 113.79 110.054 109.237 98.803 96.2293 94.0354 90.2864 87.5424
31"""
Create a chart for countries with highest U5MR
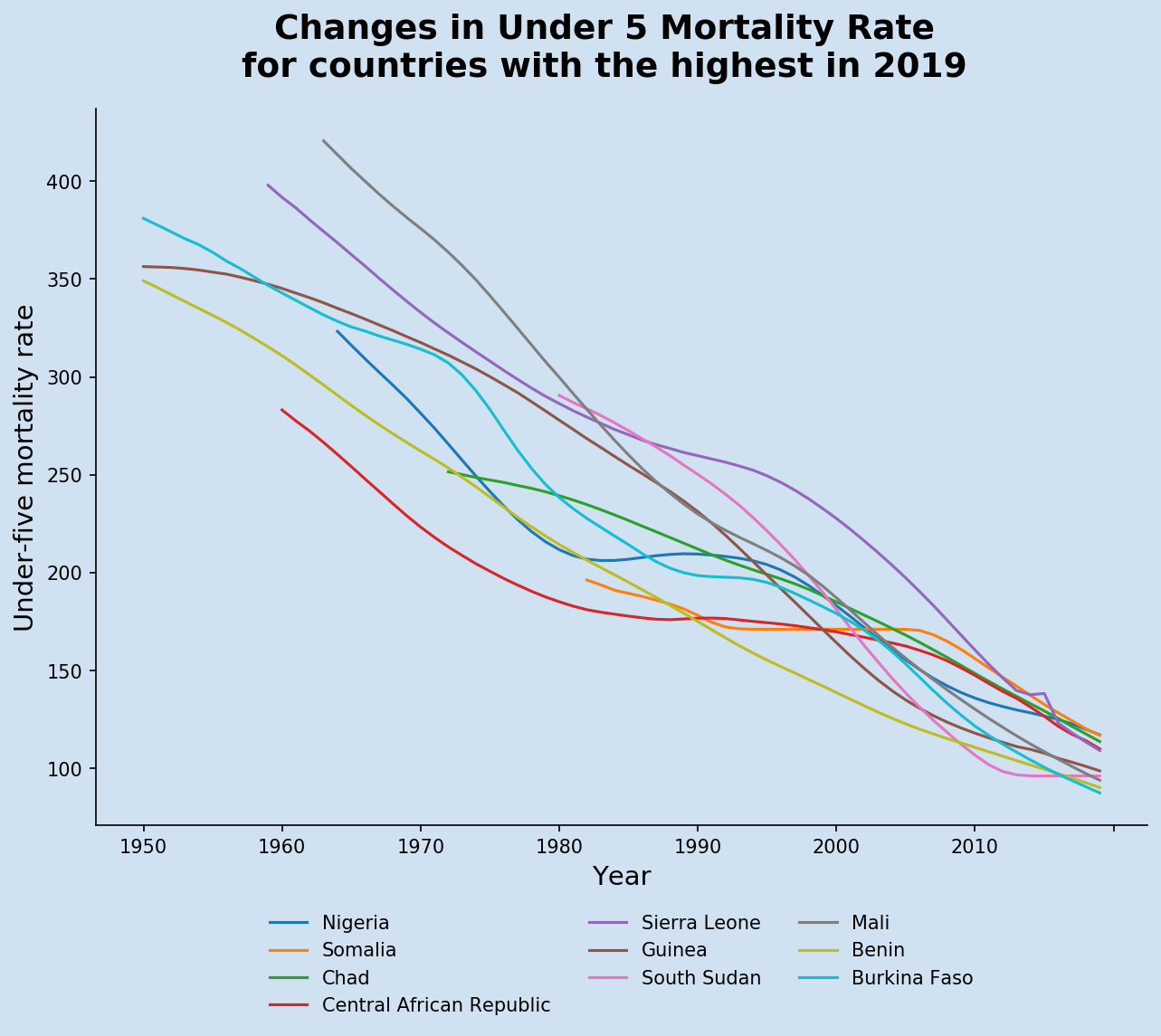
The code below creates the line chart of the changes to under-five mortality rate in the countries with the current highest rates.
1current_year = u5mr_df.columns[-1]
2title = f'Changes in Under 5 Mortality Rate\nfor countries with the highest in {u5mr_df.columns[-1]}'
3
4fig, ax1 = plt.subplots(figsize=(10,7), facecolor=plt.cm.Blues(.2))
5ax1.set_facecolor(plt.cm.Blues(.2))
6
7fig.suptitle(title, fontsize=18, fontweight='bold')
8
9top_10_df.plot(kind = "line",
10 x = "Year",
11 y = top_10_df.columns[1:],
12 ax = ax1)
13
14ax1.legend(bbox_to_anchor = (0.5, -0.1),
15 loc='upper center',
16 ncol = 3,
17 frameon = False,
18 fontsize = 'medium')
19
20ax1.set_ylabel('Under-five mortality rate', fontsize=14)
21ax1.set_xlabel('Year', fontsize=14)
22
23# Hide the right and top spines
24ax1.spines['right'].set_visible(False)
25ax1.spines['top'].set_visible(False)
26
27plt.show()
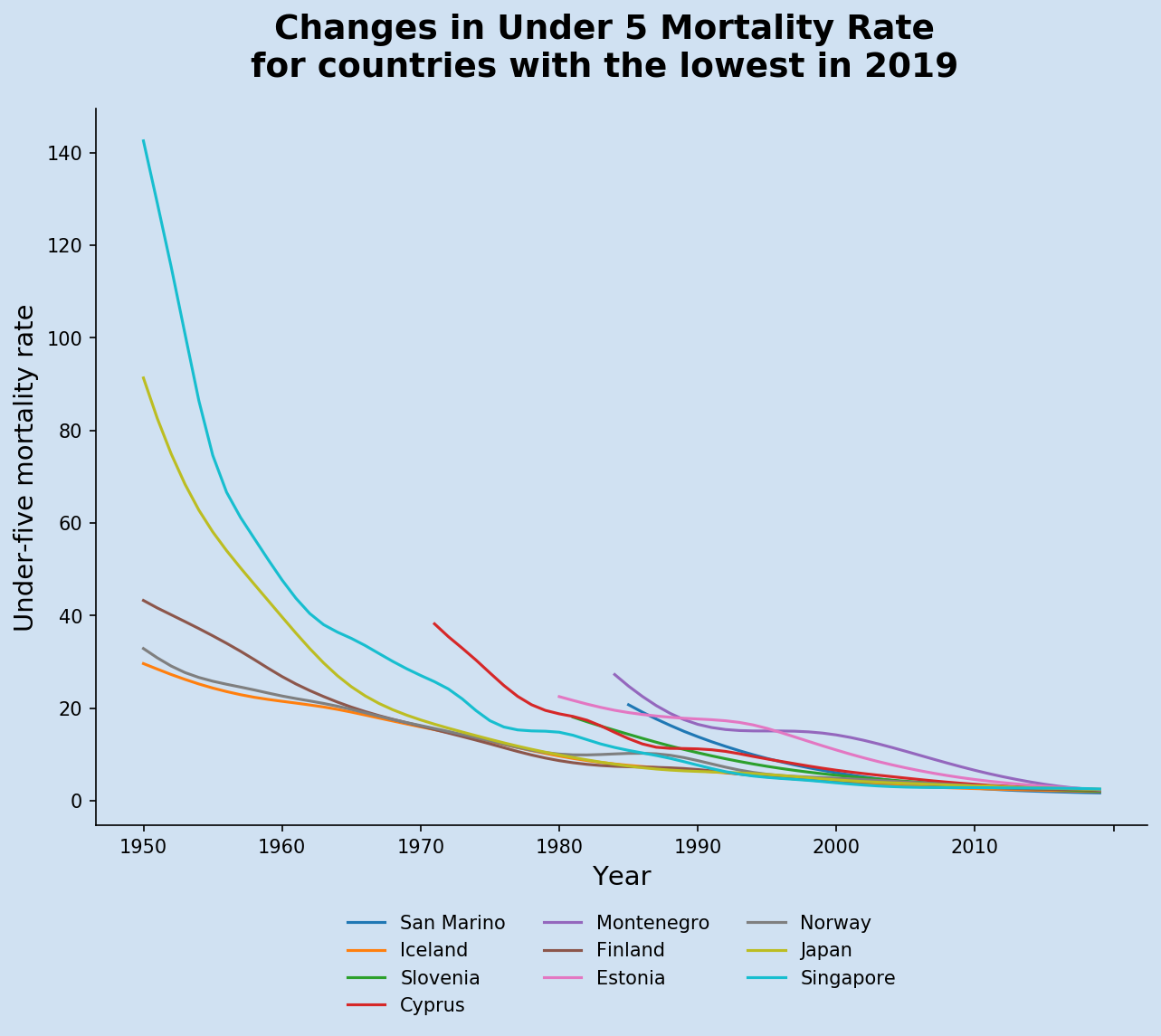
Create a chart for countries with lowest U5MR
A similar chart was created by reversing the sort order of the mortality rates in the latest year after performing the data clean. These two functions were created to make the creation of the charts more consistent and reproducible.
Function to extract the mortality rates for the top countries.
1def get_top_countries(data_df, lowest = True, num = 10):
2 '''Extract the mortality rates for the top number of countries
3 based on rates in the latest year
4
5 Keyword arguments:
6 data_df -- dateframe of all the mortality rates for all the countries
7 lowest -- boolean flag to either contries withe the lowest rate
8 or highest rate (default is True)
9 num. -- number of countries to return (default is 10)
10
11 return: dataframe that has been transposed and filtered to the top n counbtries
12 '''
13 # Need to transpose the dataframe to use year as the x-axis
14 df = data_df.sort_values(by=data_df.columns[-1], ascending=lowest).head(10).T
15 df.reset_index(drop=False,inplace=True)
16
17 # Set the Country.Name as the heading for the columns
18 df.columns = df.iloc[np.where(df['index'] == 'Country.Name')[0][0]]
19
20 # Rename the Country.Name column to Year
21 df = df.rename(columns={'Country.Name': 'Year'})
22
23 # Drop the rows that do not contain u5mr data
24 df = (df[df['Year']
25 .isin(['ISO.Code',
26 'Country.Name',
27 'Uncertainty.Bounds']) == False]
28 )
29 df.reset_index(drop = True, inplace = True)
30
31 return df
Function to create the chart of the mortality rates over time.
1def create_line_chart(df, title):
2 '''Create a line chart of the mortality rates over time.
3 Year is required to be in the first column.
4
5 Keyword arguments:
6 df -- dataframe of the the countries to render in the chart.
7 title -- text to display for the title of the chart
8
9 return: figure of the chart
10 '''
11 fig, ax1 = plt.subplots(figsize=(10,7), facecolor=plt.cm.Blues(.2))
12 ax1.set_facecolor(plt.cm.Blues(.2))
13
14 fig.suptitle(title, fontsize=18, fontweight='bold')
15
16 df.plot(kind = "line",
17 x = "Year",
18 y = df.columns[1:],
19 ax = ax1)
20
21 ax1.legend(bbox_to_anchor = (0.5, -0.1),
22 loc='upper center',
23 ncol = 3,
24 frameon = False,
25 fontsize = 'medium')
26
27 ax1.set_ylabel('Under-five mortality rate', fontsize=14)
28 ax1.set_xlabel('Year', fontsize=14)
29
30 # Hide the right and top spines
31 ax1.spines['right'].set_visible(False)
32 ax1.spines['top'].set_visible(False)
33
34 return fig
Conclusion
Under-Five Mortality Rate
Under-five mortality rate is one of the key development metrics for countries. The results of the current countries with highest continue to decrease year over year. However, some countries such as South Sudan and Somalia seem to have levelled off in recent years. These top ten countries all have U5MR close to 100, that is the expectation that 100 out of every 1,000 live births will die before the age of 5-years of age. This means one in ten people die before the age of five - this is alarming.
All of these ten countries have had high Under-Five Mortality Rate for the last fifty years and they are all improving. The highest rate in this chart is for Mali in 1963 when the Under-Five Mortality Rate was 420. This is an incomprehensible number implying that four out of every ten children are likely to die before the age of 5.
It can be seen that all but one of the top countries with the lowest Under-Five Mortality Rate started at less than 100 in 1950.
Pandas
Pandas is great for loading and manipulating structured data. It is very easy to load data from Excel into a dataframe. Dataframe is also great for filtering, sorting and transposing data as well as creating charts from the data. Pandas can be used in a Jupyter Notebook for interactive exploration of data so that a data processing pipeline can be quickly developed. The final methods can then be used in a python script to repeatedly process similar files and produce consistent output.